Computational Radiobiology
Dr. Mario A. Bernal
Associate Professor
Department of Applied Physics
“Gleb Wataghin’ Institute of Physics.
Introduction
Ionizing radiations induce biological damage in living beings. This damage originates from the impact of ionizing particles and the associated radiolysis chemical species on the DNA. Most of these damages are repaired by cellular enzymatic mechanisms. The probability for physico-chemical damage induction depends on the radiation quality and the conformation of the genetic material. Thus, each particle-energy combination shows a determined DNA damage capacity so a relative biological effectiveness may be assigned to it. For instance, heavy charged particles, like protons and alpha-particles, induce more severe DNA damages than photons for the same absorbed radiation dose.
During the last decade, the use of proton beams for radiation therapy of cancer has increaser vertiginously. This is due to the dosimetric advantages of proton over photon beams and the lowering of proton facility installation costs. However, the radiobiology associated to heavy charged particles is not well understood yet, which is an area of intense research nowadays.
Prof. Bernal has been working on this problem for more than 15 years. Two main approaches have been used so far. In the first one, Monte Carlo simulations of radiation transport is combined with geometrical and biophysical models so that damage probabilities can be determined. In the second one, more recent, quantum and classical molecular dynamics calculations are used to study the radiation-DNA collision process. Both approaches are complementary.
Research lines
1- Development of geometrical and biophysical models for estimating DNA damage yields due to the impact of ionizing radiations.
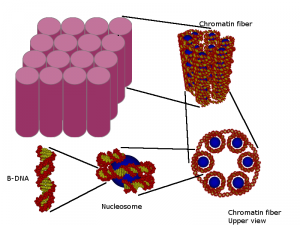
Fig. 1 Geometrical DNA model with atomic resolution developed in our group (Bernal et al. Comp. Phys. Comm. 2013).
2- Study of the proton-DNA collision using quantum and classical molecular dynamics.
Animation 1. Evolution of a DNA base-pair after the impact of a 4 keV proton.
Computational Resources.
- Planck cluster: This is the cluster of the High Performance Calculation Center in our institute. It has 4 queues with 120 cores, and several other with 36 or 12 cores each one.
- Group’s cluster: 108-Xeon cores.
Current Collaborators
- Adri van Duin. Department of Mechanical and Nuclear Engineering. Penn State University. USA.
- Angel Rubio. Director of the Theory Department of the Max Planck Institute for the Structure and Dynamics of Matter. Germany.
- Nadia Falzone. Department of Oncology. Oxford University. UK.
- Ricardo Paupitz. São Paulo State University. Brazil
- Sebastien Incerti. Research Director of the Centre National de la Recherche Scientifique. Leader of the Geant4-DNA collaboration. France
- Umberto de Giovannini. Theory Department of the Max Planck Institute for the Structure and Dynamics of Matter. Germany.
- Ziad Francis. Saint Joseph University. Lebanon.
